An up-to-date list is available on Google Scholar.
2023
-
A Robust and Efficient Framework for Slice-to-Volume Reconstruction: Application to Fetal MRI
Junshen Xu
Massachusetts Institute of Technology, 2023
Volumetric reconstruction in presence of motion is a challenging problem in medical imaging. When imaging moving targets, many modalities are limited to fast 2D imaging techniques that provide cross-sectional snapshots (2D images) of the subject with an attempt to "freeze" in-plane motion. However, inter-slice movement results in slice misalignment in 3D space, i.e., each image being an independent slice that fails to form a coherent volume for diagnosis and analysis. To this end, slice-to-volume reconstruction (SVR) has been proposed to reconstruct a high-quality 3D volume from misaligned 2D observations by performing inter-slice motion correction and super-resolution reconstruction. Existing SVR algorithms, however, have a limited capture range of slice motion and are time-consuming, particularly when producing high-resolution volumes. This thesis proposes a motion-robust and efficient machine learning framework for SVR, motivated by the application of magnetic resonance imaging (MRI) in assessing fetal brain development. We first introduce a slice-to-volume registration transformer that models input slices as a sequence and performs inter-slice motion correction by simultaneously predicting rigid transformations of all images in 3D space. We then reformulate the reconstruction problem using implicit neural representation, where the underlying volume is represented by a continuous function of 3D coordinates. This resolution-agnostic approach allows efficient reconstruction of high-resolution volumes. Finally, we extend this method to data that suffer from non-rigid motion by introducing an implicit motion field that captures slice-dependent deformation. These advances together enable robust and efficient 3D reconstruction and visualization in fetal MRI, benefiting diagnosis and downstream analysis. Additionally, the proposed framework has the potential for broader clinical implications in various applications that involve similar volumetric reconstruction problems.
-

×
![]()
NeSVoR: Implicit Neural Representation for Slice-to-Volume Reconstruction in MRI
IEEE Transactions on Medical Imaging, 2023
Reconstructing 3D MR volumes from multiple motion-corrupted stacks of 2D slices has shown promise in imaging of moving subjects, e.g., fetal MRI. However, existing slice-to-volume reconstruction methods are time-consuming, especially when a high-resolution volume is desired. Moreover, they are still vulnerable to severe subject motion and when image artifacts are present in acquired slices. In this work, we present NeSVoR, a resolution-agnostic slice-to-volume reconstruction method, which models the underlying volume as a continuous function of spatial coordinates with implicit neural representation. To improve robustness to subject motion and other image artifacts, we adopt a continuous and comprehensive slice acquisition model that takes into account rigid inter-slice motion, point spread function, and bias fields. NeSVoR also estimates pixel-wise and slice-wise variances of image noise and enables removal of outliers during reconstruction and visualization of uncertainty. Extensive experiments are performed on both simulated and in vivo data to evaluate the proposed method. Results show that NeSVoR achieves state-of-the-art reconstruction quality while providing two to ten-fold acceleration in reconstruction times over the state-of-the-art algorithms.
-
Latent Signal Models: Learning Compact Representations of Signal Evolution for Improved Time-Resolved, Multi-Contrast MRI
Magnetic Resonance in Medicine, 2023
Purpose: Training auto-encoders on simulated signal evolution and inserting the decoder into the forward model improves reconstructions through more compact, Bloch-equation-based representations of signal in comparison to linear subspaces. Methods: Building on model-based nonlinear and linear subspace techniques that enable reconstruction of signal dynamics, we train auto-encoders on dictionaries of simulated signal evolution to learn more compact, non-linear, latent representations. The proposed Latent Signal Model framework inserts the decoder portion of the auto-encoder into the forward model and directly reconstructs the latent representation. Latent Signal Models essentially serve as a proxy for fast and feasible differentiation through the Bloch-equations used to simulate signal. This work performs experiments in the context of T2-shuffling, gradient echo EPTI, and MPRAGE-shuffling. We compare how efficiently auto-encoders represent signal evolution in comparison to linear subspaces. Simulation and in-vivo experiments then evaluate if reducing degrees of freedom by inserting the decoder into the forward model improves reconstructions in comparison to subspace constraints. Results: An auto-encoder with one real latent variable represents FSE, EPTI, and MPRAGE signal evolution as well as linear subspaces characterized by four basis vectors. In simulated/in-vivo T2-shuffling and in-vivo EPTI experiments, the proposed framework achieves consistent quantitative NRMSE and qualitative improvement over linear approaches. From qualitative evaluation, the proposed approach yields images with reduced blurring and noise amplification in MPRAGE shuffling experiments. Conclusion: Directly solving for non-linear latent representations of signal evolution improves time-resolved MRI reconstructions through reduced degrees of freedom.
-
Zero-Shot Self-Supervised Joint Temporal Image and Sensitivity Map Reconstruction via Linear Latent Space
In Medical Imaging with Deep Learning – MIDL 2023
Fast spin-echo (FSE) pulse sequences for Magnetic Resonance Imaging (MRI) offer important imaging contrast in clinically feasible scan times. T2-shuffling is widely used to resolve temporal signal dynamics in FSE acquisitions by exploiting temporal correlations via linear latent space and a predefined regularizer. However, predefined regularizers fail to exploit the incoherence especially for 2D acquisitions.Recent self-supervised learning methods achieve high-fidelity reconstructions by learning a regularizer from undersampled data without a standard supervised training data set. In this work, we propose a novel approach that utilizes a self supervised learning framework to learn a regularizer constrained on a linear latent space which improves time-resolved FSE images reconstruction quality. Additionally, in regimes without groundtruth sensitivity maps, we propose joint estimation of coil-sensitivity maps using an iterative reconstruction technique. Our technique functions is in a zero-shot fashion, as it only utilizes data from a single scan of highly undersampled time series images. We perform experiments on simulated and retrospective in-vivo data to evaluate the performance of the proposed zero-shot learning method for temporal FSE reconstruction. The results demonstrate the success of our proposed method where NMSE and SSIM are significantly increased and the artifacts are reduced.
2022
-
×
![]()
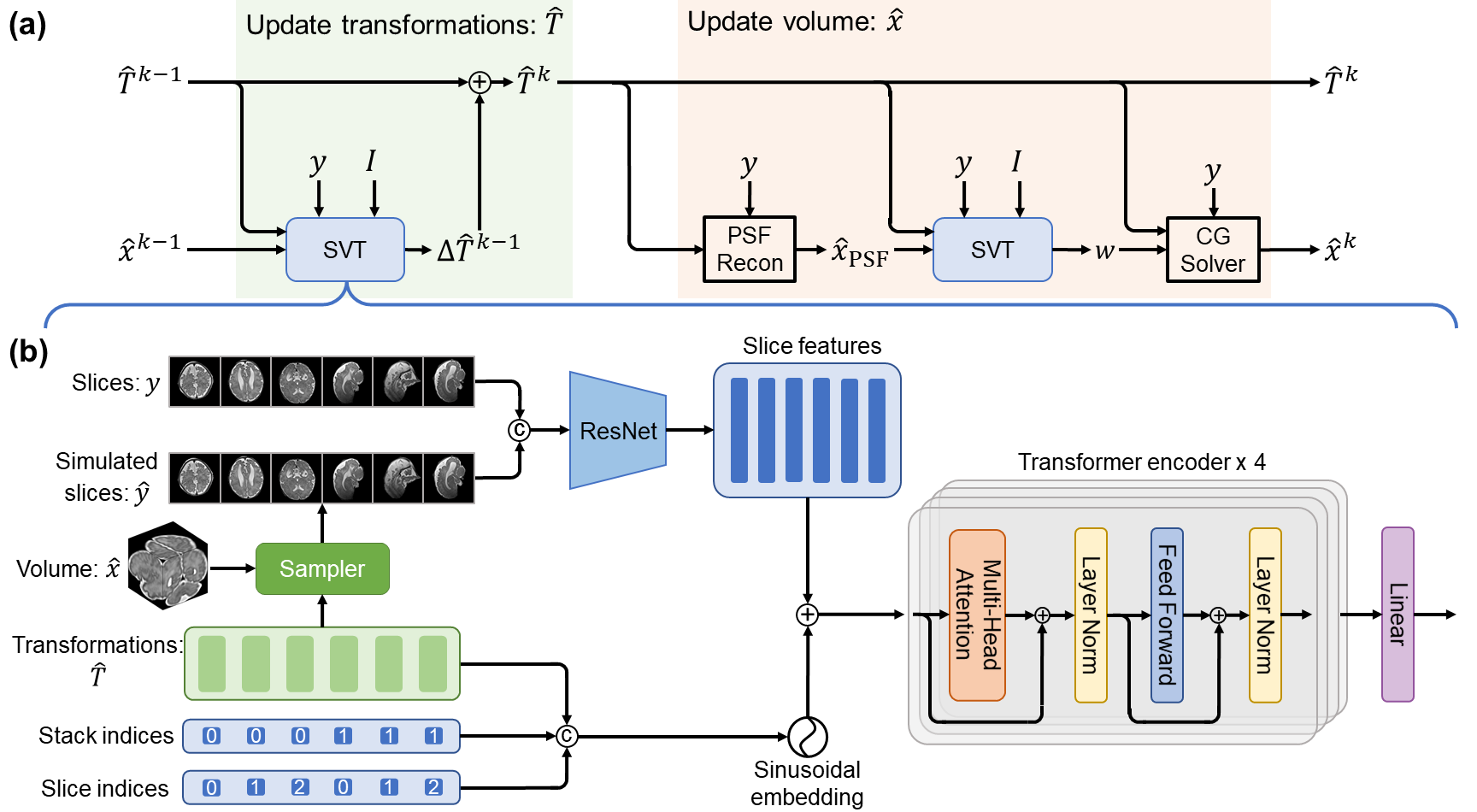
SVoRT: Iterative Transformer for Slice-to-Volume Registration in Fetal Brain MRI
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2022
Volumetric reconstruction of fetal brains from multiple stacks of MR slices, acquired in the presence of almost unpredictable and often severe subject motion, is a challenging task that is highly sensitive to the initialization of slice-to-volume transformations. We propose a novel slice-to-volume registration method using Transformers trained on synthetically transformed data, which model multiple stacks of MR slices as a sequence. With the attention mechanism, our model automatically detects the relevance between slices and predicts the transformation of one slice using information from other slices. We also estimate the underlying 3D volume to assist slice-to-volume registration and update the volume and transformations alternately to improve accuracy. Results on synthetic data show that our method achieves lower registration error and better reconstruction quality compared with existing state-of-the-art methods. Experiments with real-world MRI data are also performed to demonstrate the ability of the proposed model to improve the quality of 3D reconstruction under severe fetal motion.
-
×
![]()
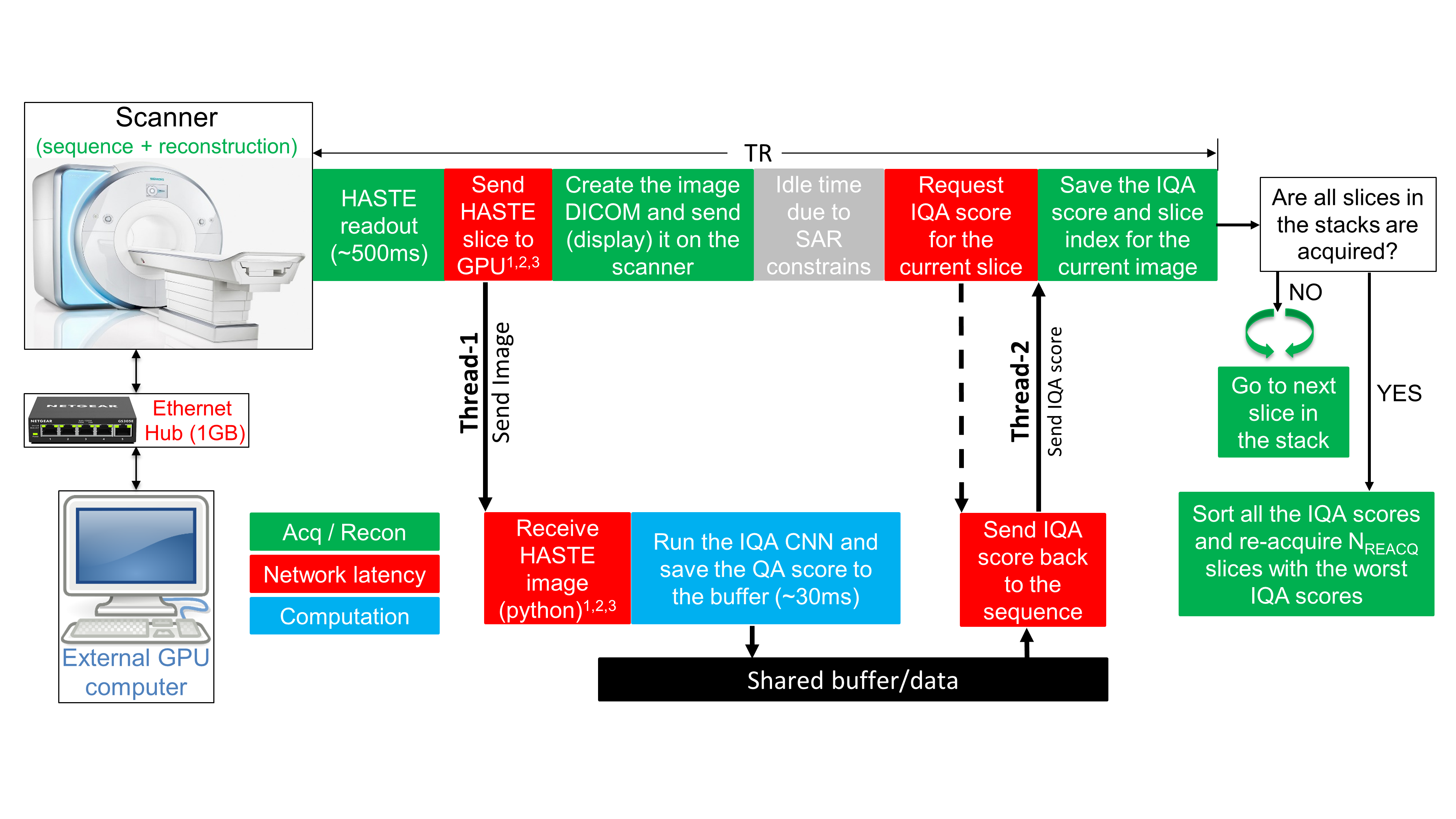
Automated Detection and Reacquisition of Motion-Degraded Images in Fetal HASTE Imaging at 3 T
Borjan Gagoski*,
Junshen Xu*, Paul Wighton, M. Dylan Tisdall, and
6 more authors
Magnetic Resonance in Medicine, 2022
Purpose: Fetal brain Magnetic Resonance Imaging suffers from unpredictable and unconstrained fetal motion that causes severe image artifacts even with half-Fourier single-shot fast spin echo (HASTE) readouts. This work presents the implementation of a closed-loop pipeline that automatically detects and reacquires HASTE images that were degraded by fetal motion without any human interaction. Methods: A convolutional neural network that performs automatic image quality assessment (IQA) was run on an external GPU-equipped computer that was connected to the internal network of the MRI scanner. The modified HASTE pulse sequence sent each image to the external computer, where the IQA convolutional neural network evaluated it, and then the IQA score was sent back to the sequence. At the end of the HASTE stack, the IQA scores from all the slices were sorted, and only slices with the lowest scores (corresponding to the slices with worst image quality) were reacquired. Results: The closed-loop HASTE acquisition framework was tested on 10 pregnant mothers, for a total of 73 acquisitions of our modified HASTE sequence. The IQA convolutional neural network, which was successfully employed by our modified sequence in real time, achieved an accuracy of 85.2% and area under the receiver operator characteristic of 0.899. Conclusion: The proposed acquisition/reconstruction pipeline was shown to successfully identify and automatically reacquire only the motion degraded fetal brain HASTE slices in the prescribed stack. This minimizes the overall time spent on HASTE acquisitions by avoiding the need to repeat the entire stack if only few slices in the stack are motion-degraded.
-
Cross-sectional Observational Study of Typical in-utero Fetal Movements using Machine Learning
Lana Vasung*, Junshen Xu*, Esra Abaci-Turk*, Cindy Zhou, and
9 more authors
Developmental Neuroscience, 2022
Early variations of fetal movements are the hallmark of a healthy developing central nervous system. However, there are no automatic methods to quantify the complex 3D motion of the developing fetus in-utero. The aim of this prospective study was to use machine learning (ML) on in-utero MRI to perform quantitative kinematic analysis of fetal limb movement, assessing the impact of maternal, placental, and fetal factors. In this cross-sectional, observational study, we used 76 sets of fetal (24-40 gestational weeks (GW)) blood oxygenation level-dependent (BOLD) MRI scans of 52 women (18-45 years old) during typical pregnancies. Pregnant women were scanned for 5 to 10 minutes while breathing room air (21% O2) and for 5 to 10 minutes while breathing 100% FiO2 in supine and/or lateral position. BOLD acquisition time was 20 minutes in total with an effective temporal resolution of approximately 3 seconds. To quantify upper and lower limb kinematics, we used a 3D convolutional neural network (CNN) previously trained to track fetal key points (wrists, elbows, shoulders, ankles, knees, hips) on similar BOLD time series. Tracking was visually assessed, errors manually corrected and the absolute movement time (AMT) for each joint was calculated. To identify variables that had a significant association with AMT, we constructed a mixed-model ANOVA with interaction terms. Fetuses showed significantly longer duration of limb movements during maternal hyperoxia. We also found a significant centrifugal increase of AMT across limbs and significantly longer AMT of upper extremities < 31 GW and longer AMT of lower extremities > 35 GW. In conclusion, using ML we successfully quantified complex 3D fetal limb motion in-utero and across gestation, showing maternal factors (hyperoxia) and fetal factors (gestational age, joint) impact movement. Quantification of fetal motion on MRI is a potential new biomarker of fetal health and neuromuscular development.
2021
-
×
![]()
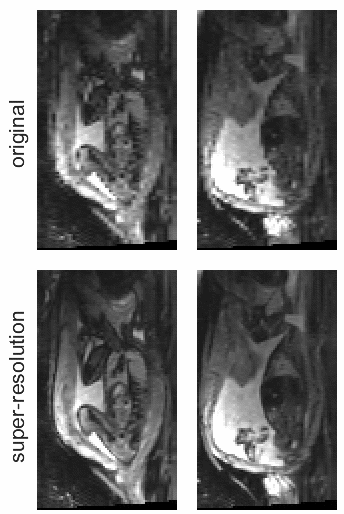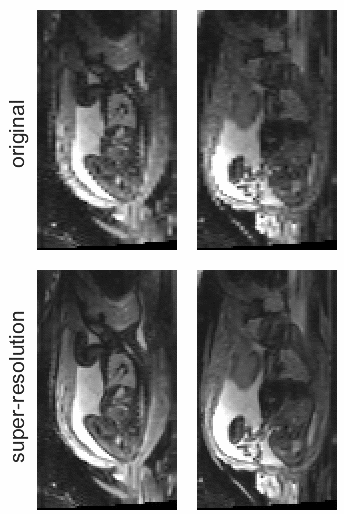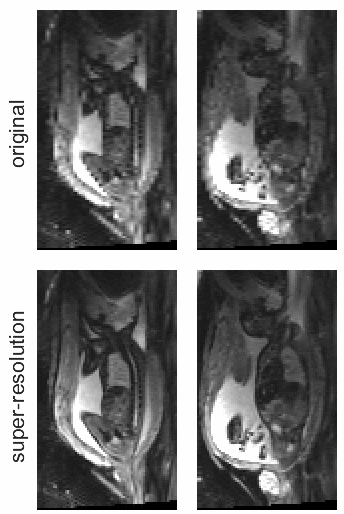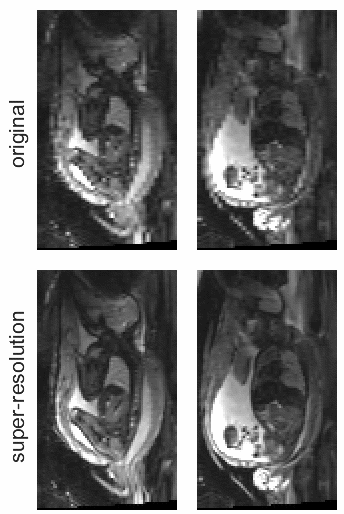
STRESS: Super-Resolution for Dynamic Fetal MRI Using Self-Supervised Learning
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021
Fetal motion is unpredictable and rapid on the scale of conventional MR scan times. Therefore, dynamic fetal MRI, which aims at capturing fetal motion and dynamics of fetal function, is limited to fast imaging techniques with compromises in image quality and resolution. Super-resolution for dynamic fetal MRI is still a challenge, especially when multi-oriented stacks of image slices for oversampling are not available and high temporal resolution for recording the dynamics of the fetus or placenta is desired. Further, fetal motion makes it difficult to acquire high-resolution images for supervised learning methods. To address this problem, in this work, we propose STRESS (Spatio-Temporal Resolution Enhancement with Simulated Scans), a self-supervised super-resolution framework for dynamic fetal MRI with interleaved slice acquisitions. Our proposed method simulates an interleaved slice acquisition along the high-resolution axis on the originally acquired data to generate pairs of low- and high-resolution images. Then, it trains a super-resolution network by exploiting both spatial and temporal correlations in the MR time series, which is used to enhance the resolution of the original data. Evaluations on both simulated and in utero data show that our proposed method outperforms other self-supervised super-resolution methods and improves image quality, which is beneficial to other downstream tasks and evaluations.
-
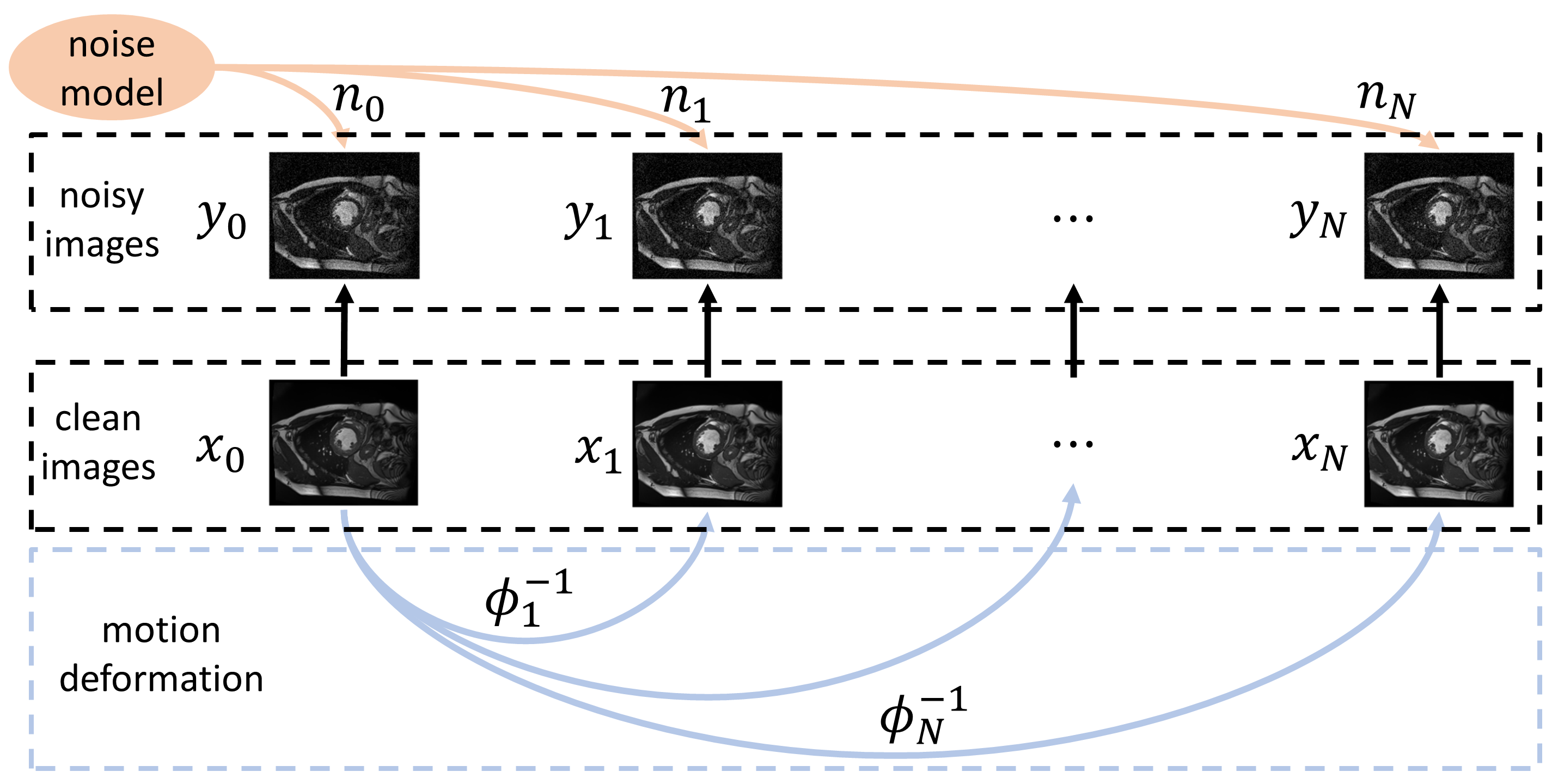
Deformed2Self: Self-Supervised Denoising for Dynamic Medical Imaging
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021
Image denoising is of great importance for medical imaging system, since it can improve image quality for disease diagnosis and downstream image analyses. In a variety of applications, dynamic imaging techniques are utilized to capture the time-varying features of the subject, where multiple images are acquired for the same subject at different time points. Although signal-to-noise ratio of each time frame is usually limited by the short acquisition time, the correlation among different time frames can be exploited to improve denoising results with shared information across time frames. With the success of neural networks in computer vision, supervised deep learning methods show prominent performance in single-image denoising, which rely on large datasets with clean-vs-noisy image pairs. Recently, several self-supervised deep denoising models have been proposed, achieving promising results without needing the pairwise ground truth of clean images. In the field of multi-image denoising, however, very few works have been done on extracting correlated information from multiple slices for denoising using self-supervised deep learning methods. In this work, we propose Deformed2Self, an end-to-end self-supervised deep learning framework for dynamic imaging denoising. It combines single-image and multi-image denoising to improve image quality and use a spatial transformer network to model motion between different slices. Further, it only requires a single noisy image with a few auxiliary observations at different time frames for training and inference. Evaluations on phantom and in vivo data with different noise statistics show that our method has comparable performance to other state-of-the-art unsupervised or self-supervised denoising methods and outperforms under high noise levels.
-
×
![]()
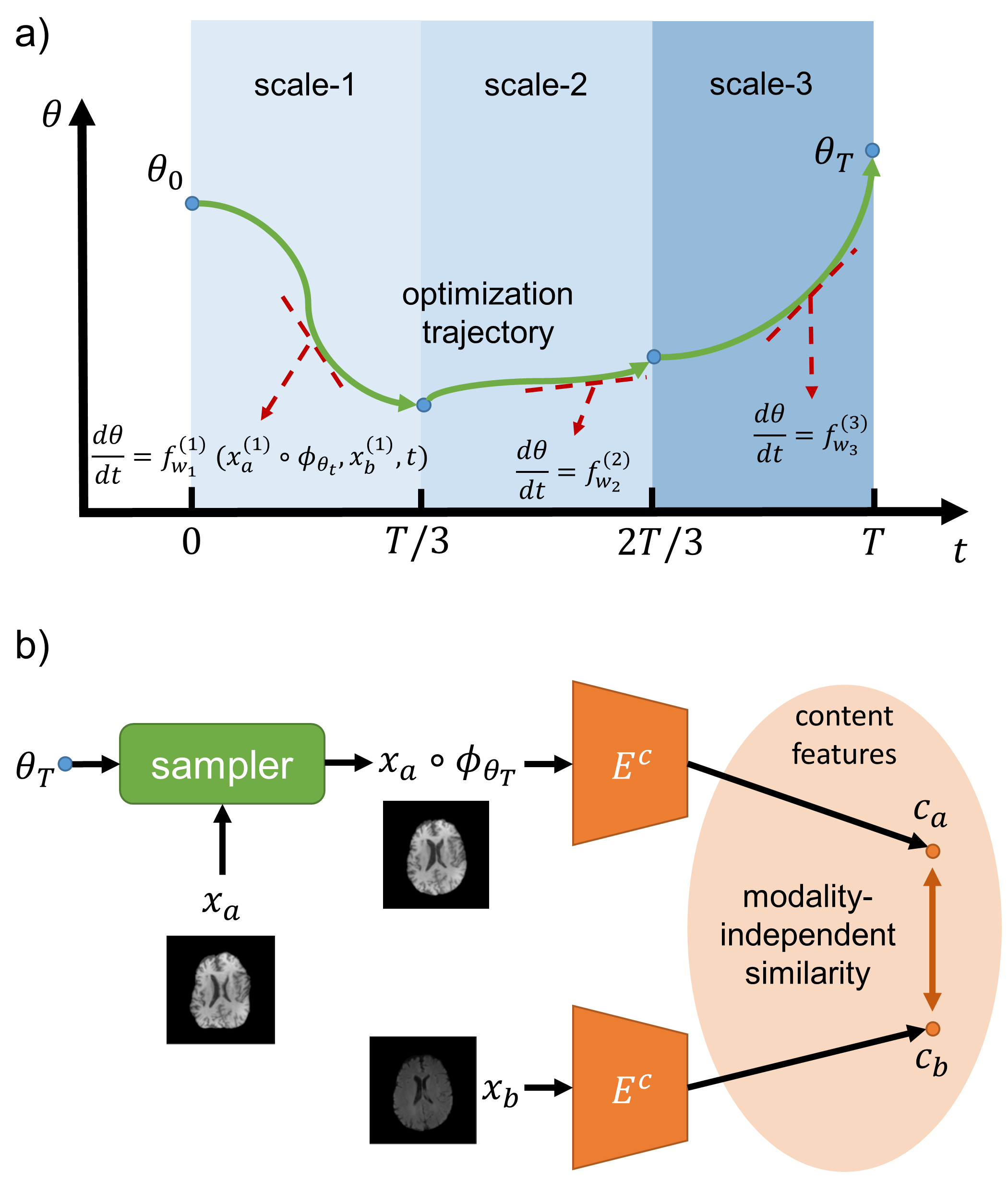
Multi-Scale Neural ODEs for 3D Medical Image Registration
Junshen Xu, Eric Z. Chen, Xiao Chen, Terrence Chen, and
1 more author
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021
Image registration plays an important role in medical image analysis. Conventional optimization based methods provide an accurate estimation due to the iterative process at the cost of expensive computation. Deep learning methods such as learn-to-map are much faster but either iterative or coarse-to-fine approach is required to improve accuracy for handling large motions. In this work, we proposed to learn a registration optimizer via a multi-scale neural ODE model. The inference consists of iterative gradient updates similar to a conventional gradient descent optimizer but in a much faster way, because the neural ODE learns from the training data to adapt the gradient efficiently at each iteration. Furthermore, we proposed to learn a modal-independent similarity metric to address image appearance variations across different image contrasts. We performed evaluations through extensive experiments in the context of multi-contrast 3D MR images from both public and private data sources and demonstrate the superior performance of our proposed methods.
2020
-
Online, Low-Latency Decision Making for Fetal Magnetic Resonance Imaging with Machine Learning
Junshen Xu
Massachusetts Institute of Technology, 2020
Fetal Magnetic Resonance Imaging (MRI) with T2-weighted Half-Fourier-Acquisition Single-Shot Turbo-Spin-Echo (HASTE) sequence plays an important role in diagnosing brain abnormality. However, the quality of HASTE images routinely suffer from fetal motion which leads to image artifacts, incomplete brain coverage as well as longer scan times. To address this problem, interleaved 3D Echo-planar Imaging (EPI) navigators are acquired along with HASTE images, which can provide pose information for prospective motion correction. In this thesis, we first propose a fetal pose estimation model which detects important fetal landmarks from 3D EPI data using a deep convolution neural network. We further demonstrate its capability by applying this model to fetal motion analysis. In an attempt to improve the current fetal MRI protocol, we develop a machine learning based online decision making system for fetal MRI to improve the efficiency of acquiring high quality HASTE images for clinical diagnosis. The proposed system leverages an Image Quality Assessment (IQA) network to determine whether an acquired HASTE slice is contaminated by motion artifacts and improves image quality by re-acquisition. Evaluation on retrospective experiments and in vivo scans suggests that the proposed pipeline can improve image quality with a reasonable number of re-acquisition, potentially enabling a more efficient workflow for fetal brain MRI.
-
×
![]()
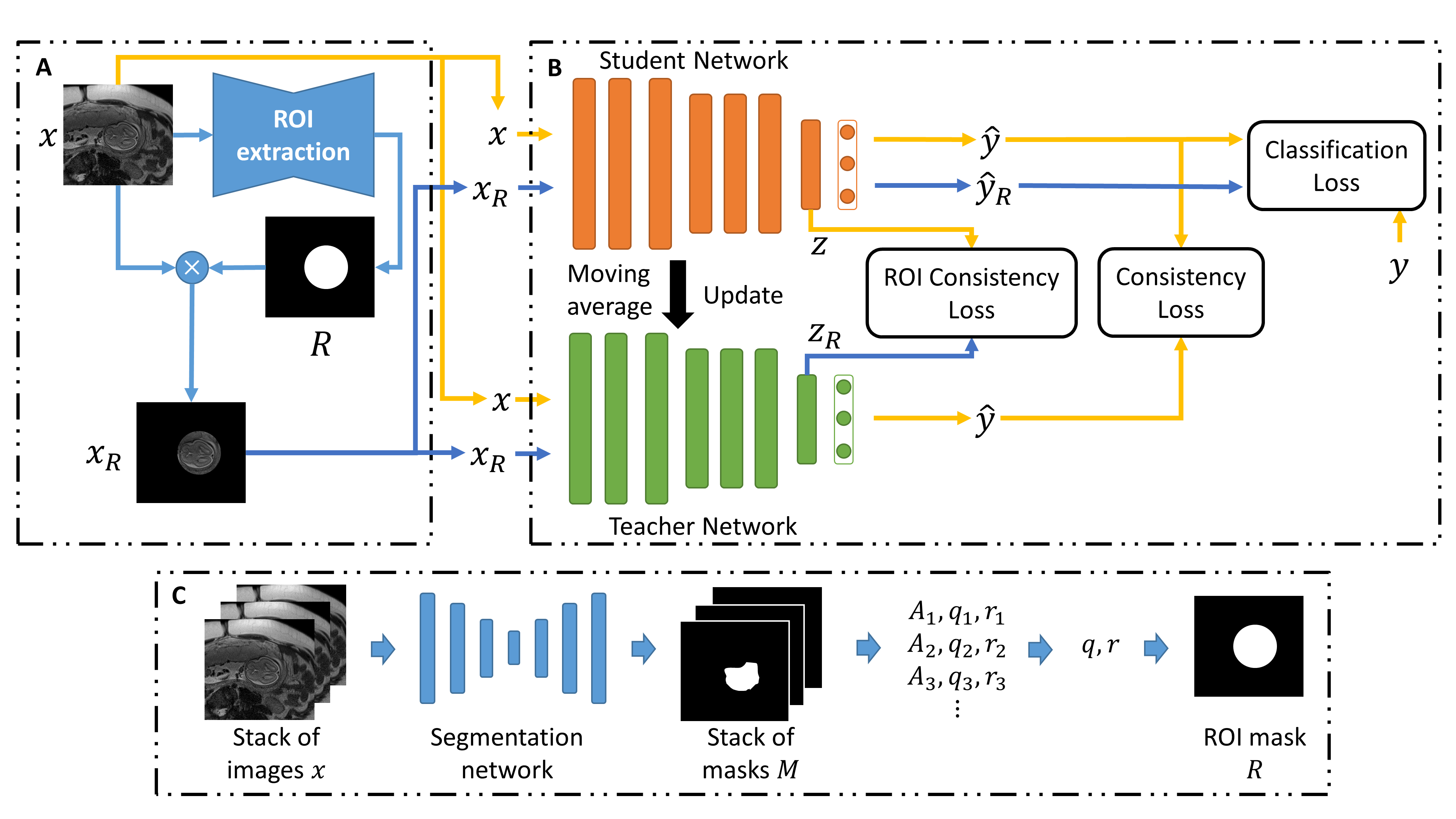
Semi-Supervised Learning for Fetal Brain MRI Quality Assessment with ROI Consistency
Junshen Xu, Sayeri Lala,
Borjan Gagoski, Esra Abaci Turk, and
3 more authors
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020
Fetal brain MRI is useful for diagnosing brain abnormalities but is challenged by fetal motion. The current protocol for T2-weighted fetal brain MRI is not robust to motion so image volumes are degraded by inter- and intra- slice motion artifacts. Besides, manual annotation for fetal MR image quality assessment are usually time-consuming. Therefore, in this work, a semi-supervised deep learning method that detects slices with artifacts during the brain volume scan is proposed. Our method is based on the mean teacher model, where we not only enforce consistency between student and teacher models on the whole image, but also adopt an ROI consistency loss to guide the network to focus on the brain region. The proposed method is evaluated on a fetal brain MR dataset with 11,223 labeled images and more than 200,000 unlabeled images. Results show that compared with supervised learning, the proposed method can improve model accuracy by about 6% and outperform other state-of-the-art semi-supervised learning methods. The proposed method is also implemented and evaluated on an MR scanner, which demonstrates the feasibility of online image quality assessment and image reacquisition during fetal MR scans.
-
Enhanced Detection of Fetal Pose in 3D MRI by Deep Reinforcement Learning with Physical Structure Priors on Anatomy
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020
Fetal MRI is heavily constrained by unpredictable and substantial fetal motion that causes image artifacts and limits the set of viable diagnostic image contrasts. Current mitigation of motion artifacts is predominantly performed by fast, single-shot MRI and retrospective motion correction. Estimation of fetal pose in real time during MRI stands to benefit prospective methods to detect and mitigate fetal motion artifacts where inferred fetal motion is combined with online slice prescription with low-latency decision making. Current developments of deep reinforcement learning (DRL), offer a novel approach for fetal landmarks detection. In this task 15 agents are deployed to detect 15 landmarks simultaneously by DRL. The optimization is challenging, and here we propose an improved DRL that incorporates priors on physical structure of the fetal body. First, we use graph communication layers to improve the communication among agents based on a graph where each node represents a fetal-body landmark. Further, additional reward based on the distance between agents and physical structures such as the fetal limbs is used to fully exploit physical structure. Evaluation of this method on a repository of 3-mm resolution in vivo data demonstrates a mean accuracy of landmark estimation 10 mm of ground truth as 87.3%, and a mean error of 6.9 mm. The proposed DRL for fetal pose landmark search demonstrates a potential clinical utility for online detection of fetal motion that guides real-time mitigation of motion artifacts as well as health diagnosis during MRI of the pregnant mother.
-
3D Fetal Pose Estimation with Adaptive Variance and Conditional Generative Adversarial Network
In Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis, 2020
Fetal motion is the dominant challenge to reliable performance and diagnostic quality of fetal magnetic resonance imaging (MRI). The fetus can move unpredictably and rapidly, leading to severe image artifacts. Consequently, MR acquisitions are largely limited to so-called single-shot techniques in an attempt to “freeze” fetal motion through fast imaging, while the problem due to motion occur between slices still exists. In this work, we propose a deep learning method for fetal pose estimation from MR volumes using the paradigm of conditional generative adversarial network which consists of two networks, a generator and a discriminator. The generator is responsible for estimating keypoint heatmaps from input MRI and the discriminator tries to learn the features of plausible fetal pose and distinguish ground-truth heatmaps from generated ones. With this adversarial training scheme, the generator can robustly produce realistic heatmaps for fetal pose inference. Besides, we use adaptive variance to model the difference in intensity of motion of different keypoints. Evaluation shows that the proposed method can improve the performance of pose estimation in 3D MRI, achieving quantitatively an average error of 2.64 mm and 98.31% accuracy (with error less than 10 mm). The proposed method can process volumes with latency less than 300 ms, potentially enabling low-latency online tracking of fetal pose during MR scans.
-
×
![]()
Ultra-Low-Dose 18F-FDG Brain PET/MR Denoising using Deep Learning and Multi-Contrast Information
Junshen Xu, Enhao Gong, Jiahong Ouyang,
John Pauly, and
1 more author
In Medical Imaging 2020: Image Processing
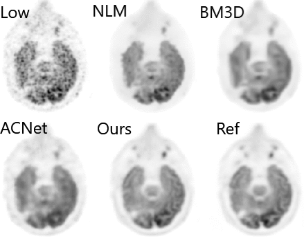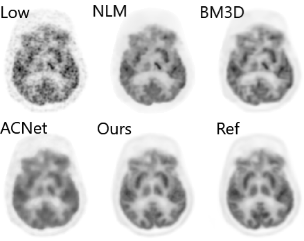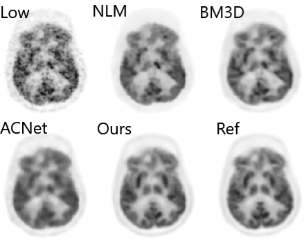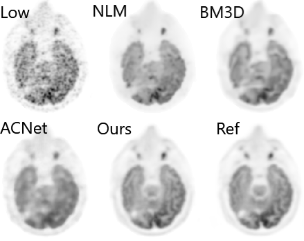
Positron emission tomography (PET) is a widely used molecular imaging modality for various clinical applications. With Magnetic Resonance Imaging (MRI) providing anatomical information, simultaneous PET/MR reduces the radiation risk. Both improved hardware and algorithms have been developed to further reduce the amount of radiotracer dosage, but these methods are not yet applied to very low dose. Here, we propose a Deep Learning based method to enable ultra-low-dose PET denoising with multi-contrast information from simultaneous MRI. Methods:The method is implemented to denoise 18F-fluorodeoxyglucose (FDG) brain PET images from low-dose images with 200-fold dose reduction through undersampling, and evaluated for glioblastoma (GBM) patients. Comprehensive quantitative and qualitative evaluations were conducted to verify the performance and clinical applicability of the proposed method, including quantitative accuracy evaluation, visual quality evaluation, reader study with manual tumor segmentation to evaluate the diagnostic quality. Results:The results demonstrate that the proposed method achieves superior results in performance and efficiency comparing with the state-of-art denoising methods. Conclusion:Though reconstructed from scans with only 0.5% of the standard dose, the denoised ultra-low-dose PET images deliver similar visual quality and diagnostic information as the standard-dose PET images. By combining PET and MR information, the proposed Deep Learning based method improves image quality of ultra-low-dose PET, preserves diagnostic quality, and potentially enables much safer, faster, and more cost-effective PET/MR studies.
2019
-
×
![]()
Fetal Pose Estimation in Volumetric MRI using a 3D Convolution Neural Network
In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019
The performance and diagnostic utility of magnetic resonance imaging (MRI) in pregnancy is fundamentally constrained by fetal motion. Motion of the fetus, which is unpredictable and rapid on the scale of conventional imaging times, limits the set of viable acquisition techniques to single-shot imaging with severe compromises in signal-to-noise ratio and diagnostic contrast, and frequently results in unacceptable image quality. Surprisingly little is known about the characteristics of fetal motion during MRI and here we propose and demonstrate methods that exploit a growing repository of MRI observations of the gravid abdomen that are acquired at low spatial resolution but relatively high temporal resolution and over long durations (10–30 min). We estimate fetal pose per frame in MRI volumes of the pregnant abdomen via deep learning algorithms that detect key fetal landmarks. Evaluation of the proposed method shows that our framework achieves quantitatively an average error of 4.47 mm and 96.4% accuracy (with error less than 10 mm). Fetal pose estimation in MRI time series yields novel means of quantifying fetal movements in health and disease, and enables the learning of kinematic models that may enhance prospective mitigation of fetal motion artifacts during MRI acquisition.
-
Ultra-Low-Dose 18F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs
Kevin T. Chen, Enhao Gong, Fabiola Bezerra Carvalho Macruz, Junshen Xu, and
9 more authors
Radiology, 2019
PMID: 30526350
PurposeTo reduce radiotracer requirements for amyloid PET/MRI without sacrificing diagnostic quality by using deep learning methods.Materials and MethodsForty data sets from 39 patients (mean age ± standard deviation [SD], 67 years ± 8), including 16 male patients and 23 female patients (mean age, 66 years ± 6 and 68 years ± 9, respectively), who underwent simultaneous amyloid (fluorine 18 [18F]–florbetaben) PET/MRI examinations were acquired from March 2016 through October 2017 and retrospectively analyzed. One hundredth of the raw list-mode PET data were randomly chosen to simulate a low-dose (1%) acquisition. Convolutional neural networks were implemented with low-dose PET and multiple MR images (PET-plus-MR model) or with low-dose PET alone (PET-only) as inputs to predict full-dose PET images. Quality of the synthesized images was evaluated while Bland-Altman plots assessed the agreement of regional standard uptake value ratios (SUVRs) between image types. Two readers scored image quality on a five-point scale (5 = excellent) and determined amyloid status (positive or negative). Statistical analyses were carried out to assess the difference of image quality metrics and reader agreement and to determine confidence intervals (CIs) for reading results.ResultsThe synthesized images (especially from the PET-plus-MR model) showed marked improvement on all quality metrics compared with the low-dose image. All PET-plus-MR images scored 3 or higher, with proportions of images rated greater than 3 similar to those for the full-dose images (−10% difference [eight of 80 readings], 95% CI: −15%, −5%). Accuracy for amyloid status was high (71 of 80 readings [89%]) and similar to intrareader reproducibility of full-dose images (73 of 80 [91%]). The PET-plus-MR model also had the smallest mean and variance for SUVR difference to full-dose images.ConclusionSimultaneously acquired MRI and ultra–low-dose PET data can be used to synthesize full-dose–like amyloid PET images.© RSNA, 2018Online supplemental material is available for this article.See also the editorial by Catana in this issue.
-
Assessing EGFR Gene Mutation Status in Non-Small Cell Lung Cancer with Imaging Features from PET/CT
Mengmeng Jiang, Yiqian Zhang, Junshen Xu, Min Ji, and
6 more authors
Nuclear medicine communications, 2019
2017
-
×
![]()
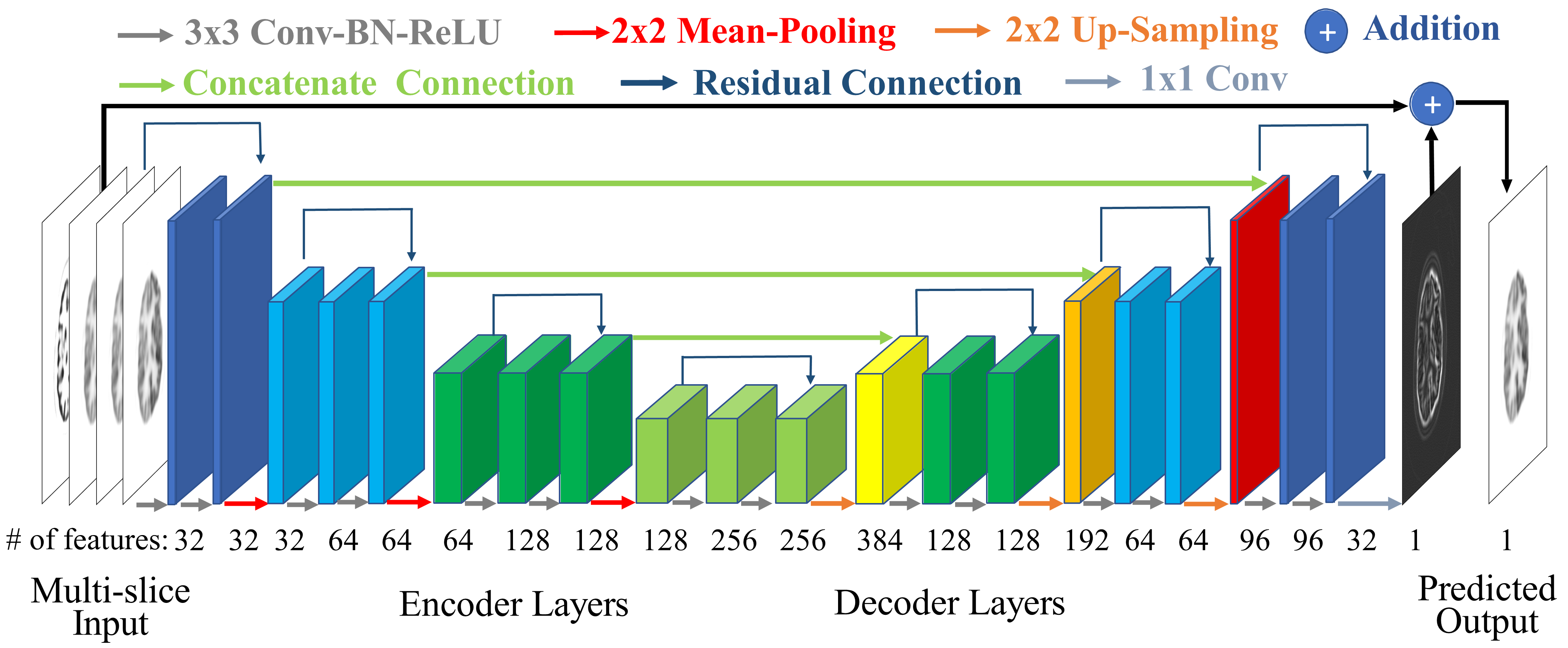
200x Low-Dose PET Reconstruction using Deep Learning
arXiv preprint arXiv:1712.04119, 2017
Positron emission tomography (PET) is widely used in various clinical applications, including cancer diagnosis, heart disease and neuro disorders. The use of radioactive tracer in PET imaging raises concerns due to the risk of radiation exposure. To minimize this potential risk in PET imaging, efforts have been made to reduce the amount of radio-tracer usage. However, lowing dose results in low Signal-to-Noise-Ratio (SNR) and loss of information, both of which will heavily affect clinical diagnosis. Besides, the ill-conditioning of low-dose PET image reconstruction makes it a difficult problem for iterative reconstruction algorithms. Previous methods proposed are typically complicated and slow, yet still cannot yield satisfactory results at significantly low dose. Here, we propose a deep learning method to resolve this issue with an encoder-decoder residual deep network with concatenate skip connections. Experiments shows the proposed method can reconstruct low-dose PET image to a standard-dose quality with only two-hundredth dose. Different cost functions for training model are explored. Multi-slice input strategy is introduced to provide the network with more structural information and make it more robust to noise. Evaluation on ultra-low-dose clinical data shows that the proposed method can achieve better result than the state-of-the-art methods and reconstruct images with comparable quality using only 0.5% of the original regular dose.